Viral infections are a significant health burden. Viruses of H1N1, HIV, Dengue, SARS and more recently COVID-19 have life threatening impacts because of their high genetic diversity, replication capability and persistence in hosts. Although vaccines and drugs are available, the virus’ constant mutations makes it necessary to continuously invent antivirals.
Antiviral peptides are short, cationic peptides composed of amino acid sequences and structures. They have the potential to be effective antiviral therapeutic agents because they can impact the entire viral lifecycle, from attachment to host cells to impairing viral replication.
However, identifying novel peptides against a specific virus family is a laborious and time-consuming process. It involves several steps such as identifying the viral family, determining the peptide backbone, finding the source, performing bioinformatics analysis, multiple assays, mutational analysis etc.
In short, discovering and synthesizing antiviral peptides is a complex multi-factor optimization problem.
Artificial intelligence-based approaches have shown promise in reducing time and improving efficiency in such processes. Machine learning algorithms can learn to identify patterns and understand data distribution of an antiviral peptide’s physico-chemical properties such as charge, aromaticity, amino acid composition, etc.
In this article, we show how a generative adversarial network (GAN) -based framework — PandoraGAN — can be used to generate potential antiviral peptides which may act against a wide variety of viruses.
What is GAN?
Generative adversarial network or GAN, for short, is a machine learning framework used to generate realistic images and text.
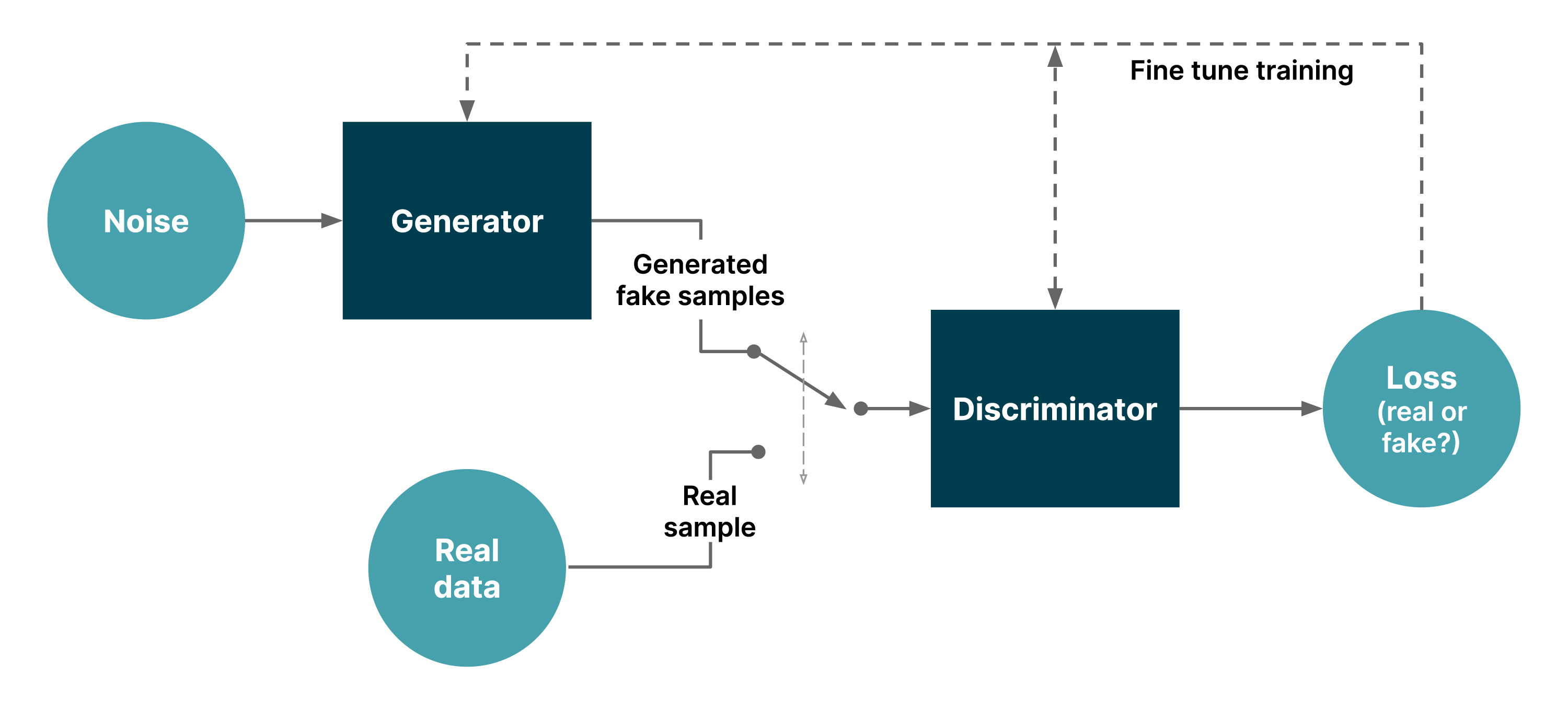

Figure 1: a simple GAN
Every GAN has a generator module and a discriminator module. The generator generates fake samples of data (be it an image, audio or a peptide) to fool the discriminator. The discriminator’s job is to distinguish between the real and fake samples. The generative model tries to capture the distribution of data and is trained to maximize the probability of the discriminator making a classification mistake. The generator and the discriminator are both neural networks, running in competition with each other in the training phase. They repeat each step several times, and get better and better with each repetition.
GANs are deep learning-based generative models that learn data distribution using gradient descent without needing prior knowledge of the structure of the data.
How do GANs work?
We need the GAN to understand ‘what makes an antiviral peptide antiviral’ based on the dataset that we feed. Ideally, the more instances (actual antiviral peptides) it comes across, the more it can learn from them and generate peptides that have antiviral properties. Typically, deep learning models require 10s to 100s of thousands of data points.
However, in our case, this was a challenge. We had 130 highly active peptides that were collected from databases and literature surveys. These were, however, high-quality, efficient, highly active peptides with a minimal inhibition concentration (MIC) of less than 15 micromolar. This makes it a good set for the underlying architecture in PandoraGAN to be able to learn a good representation of the implicit properties of antiviral peptides.
We chose LeakGAN, a variation of the vanilla GAN, because it leaks the high-level feature information from the discriminator to the generator, enabling better learning. This way, leakGAN deciphered the underlying patterns from the limited set of peptides, which helped generate potential good quality antiviral peptides.
Using PandoraGAN to generate antiviral peptides
PandoraGAN generates sequences with lengths and molecular weights similar to those in training data. However, we also need to know whether these sequences were biologically active. To evaluate this, we calculated certain physico-chemical properties like amino acid composition, dipeptide composition, residue repeat patterns, aromaticity, net charge, gravy index, secondary structure propensity, etc. for the training data, PandoraGAN output sequences and non-antiviral peptides. Based on this, we formulated rules and patterns that will validate the output sequences of PandoraGAN. The figure below shows the similarity in patterns observed for Shannon entropy for training data and PandoraGAN sequences.
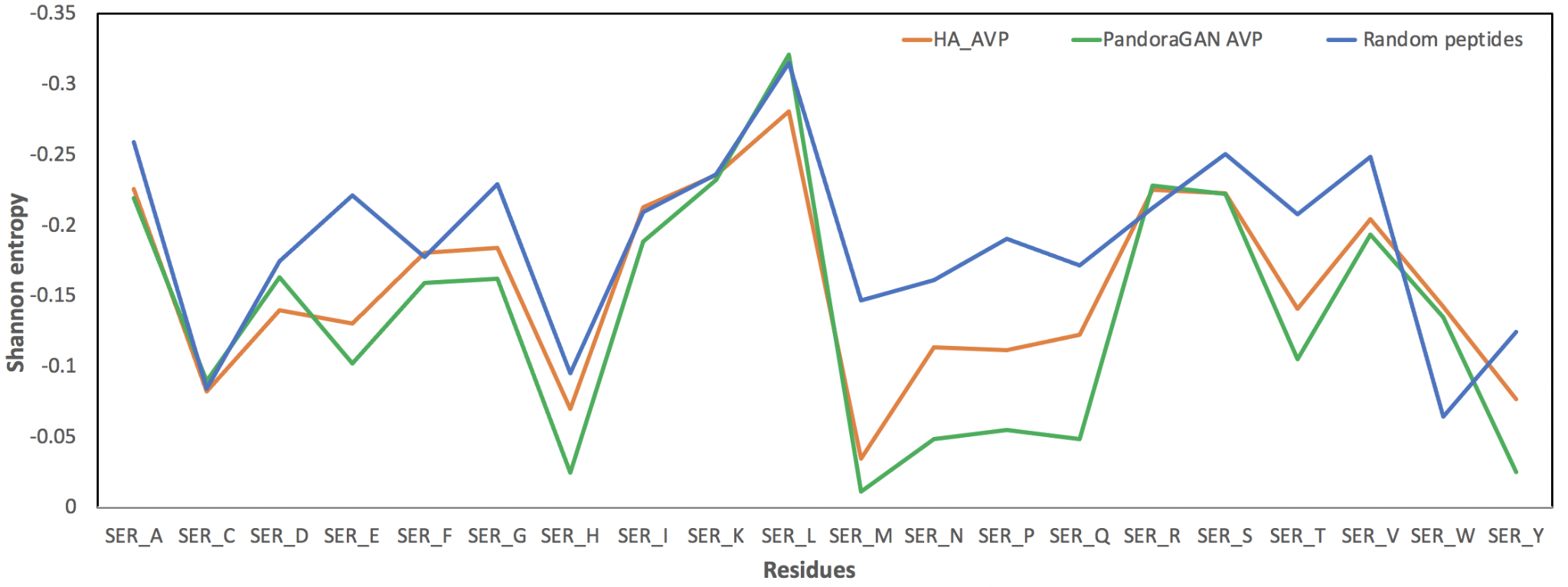

Figure 2: Shannon entropy for training data and PandoraGAN output
The Shannon entropy in the above figure shows a similar pattern for training data and PandoraGAN output as compared to the random non-antiviral peptides. For example, the peak for methionine (ser_M) confirms that PandoraGAN output is also able to generate peptides which have less methionine as seen in training data (HA_AVP).
The peaks (high entropy) and valleys (low entropy) for the Shannon entropy of residues are highly correlated with Pearson's ρ = 0.96 with p-value < 1e-10. Other physio-chemical properties such as aromaticity, instability index, isoelectric point, helix, turn, sheet, gravy and net charge at pH7.4 also follow similar distribution as the curated peptides with a Mann-Whitney U-Test p-value < 0.05. The figure below shows the overlap between training data and sequences generated by PandoraGAN in the first two components of principal component analysis (PCA).
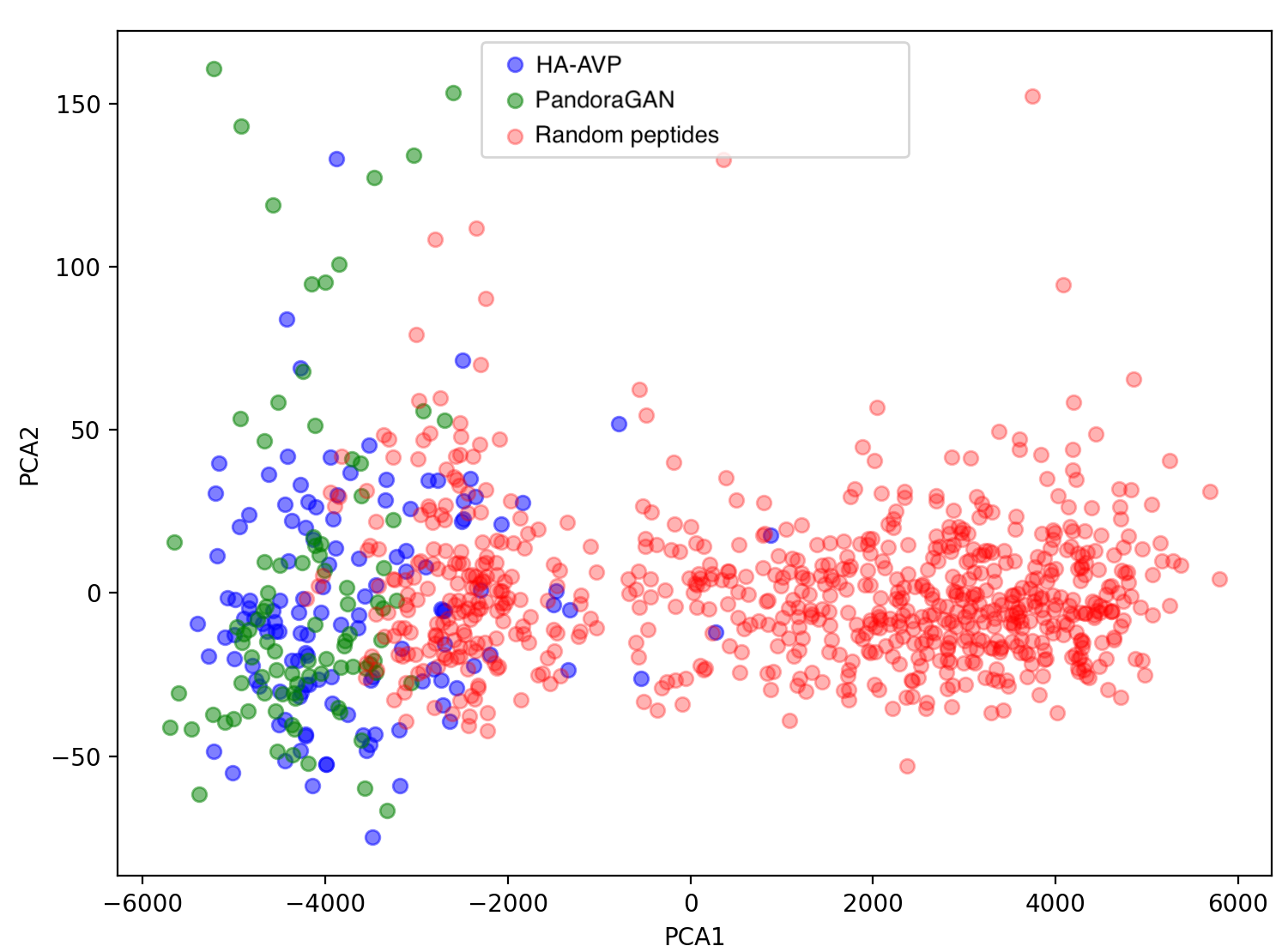

Figure 3: PCA plot of first two components
The first two components of the PCA plot show an overlap of the curated training peptides (blue) and PandoraGAN generated peptides (green). There is a clear distinction between these and the random non-secretory peptides (red).
The novelty of our approach is in generating peptides using very few data (only 130 peptides). However, since these have been manually curated, they are of quality representation, the GAN model was able to efficiently learn the distribution and properties and produce very similar peptides. The GAN based approach may create a novel variety of antiviral peptides that may not have been seen earlier and could be of therapeutic importance.
Future work and prospects
Generative methods are gaining importance in synthetic biology, where they are used to generate realistic sequences and structures. Generating antivirals using GANs can offer researchers thousands of new sequences that can be further studied in a wet lab for biological relevance and patterns. They can also be further chemically modified to increase its activity and efficacy. We believe that using PandoraGAN may significantly reduce the time and energy taken to generate peptides from scratch.
Here is the code and the paper with more details on the PandoraGAN project.
Disclaimer: The statements and opinions expressed in this article are those of the author(s) and do not necessarily reflect the positions of Thoughtworks.


















